TEAM
STÉPHANE PYRONNET & FABIENNE MEGGETTO
R’n Blood :
RNA Biology in Hematologic Tumors

The Toulouse Cancer Laboratory of Excellence is a project that aims to understand the mechanisms of resistance and relapse in cancer.
THE SPECIFICITIES
OF OUR RESEARCH AXIS
Post-transcriptional events involving RNAs allow for rapid control of gene expression at low energy cost to the cell. They are particularly solicited when cells are subjected to a stressed environment, as in a malignant tumor or under the effect of anti-tumor treatments. They can then contribute to the adaptation processes necessary for malignant transformation and/or resistance to treatments.
In a clinical perspective, using combined approaches of new generation and high throughput RNA sequencing (“bulk” or “single-cell RNAseq”) and cellular and tissue imaging, our team explores the whole RNA repertoire (coding mRNAs and non-coding regulatory RNAs including miRNAs, lncRNAs and circRNAs) and its heterogeneity within the tumor, in cohorts of patients with hematological tumors (acute myeloid leukemias and pediatric T lymphomas). The objective is the identification of new therapeutic and/or sensitizing targets.
On a cognitive and mechanistic level, the team then implements molecular (gain/loss of function), cellular and in vivo (genetic models and human tumor avatars or PDXs) experimental approaches for characterizing the new potential targets functionally and for deciphering their mechanisms of action. In partnership with industrialists, the possibility of targeting them with molecules offering a therapeutic potential is also explored.
Translation of mRNAs
Non-coding RNA
miRNA
lncRNA
circRNA
Reticulum stress
Acute leukemia
Pediatric T lymphoma
Oncogenesis
Treatment resistance
Targeted therapies
Biomarkers
RESEARCH PROJETCS
Translational control of gene expression in hematological tumors
Stéphane Pyronnet
Regulation of microarn and circular arn and their theranostic potential in alk tyrosine kinase-associated anaplastic t-cell lymphoma
Fabienne Meggetto & Laurence Lamant
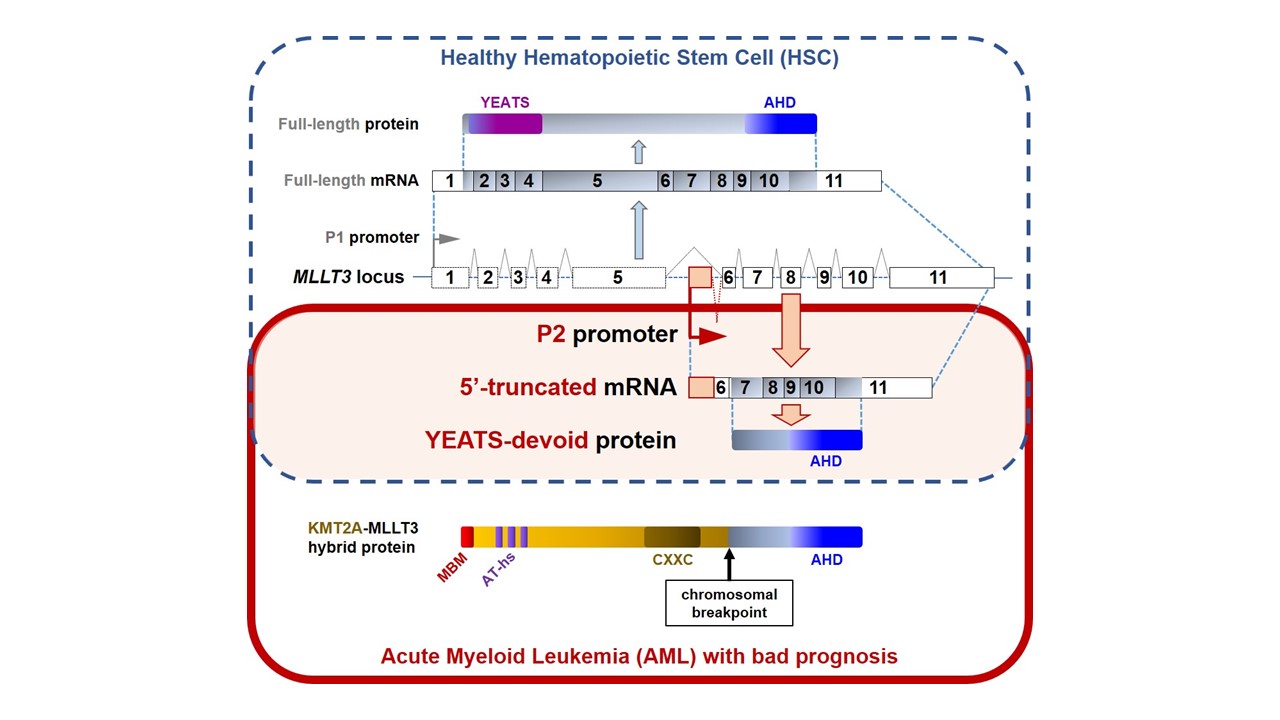
Role of endoplasmic reticulum stress in the progression and resistance of acute myeloid leukemia
Christian Touriol
Involvement of long non-coding arns in chemoresistance in acute myeloblastic leukemia
Marina Bousquet
THE TEAM’S
FOCUS
Discover
Understand
Participate
SCIENTIFIc PRODUCTIONS
PUBLICATIONS 2026
PUBLICATIONS 2025
PUBLICATIONS 2024
PUBLICATIONS 2023
PUBLICATIONS 2022
PUBLICATIONS 2021
PUBLICATIONS 2020
PUBLICATIONS 2019
team members
partnerships & FUNDING

Centre de Recherches en Cancérologie de Toulouse (Oncopole)
Toulouse – FR
Nous contacter
05 82 74 15 75
Envie de rejoindre
L’équipe du CRCT ?