Identification of therapeutic targets and repositioning of drugs against COVID-19 through the analysis of interaction networks between proteins, drugs, pathologies and symptoms
COVID-19
complex networks
interactome
Proteines
symptoms
Recent years have seen the rise of the field of network medicine, a discipline that aims to harness networks and their analysis to describe and understand the complex relationships between biological processes, drugs, phenotypes and, ultimately, diseases.
Never before has this approach been so relevant to the global medical community, in order to find a cure for a new disease that suddenly appears and rapidly takes its toll.
COVID-19, the disease caused by the SARS-CoV-2 virus infection, was officially identified in January 2020 and since then the pace of science has exceeded what we thought possible. Very quickly, patient data began to be collected and hundreds of treatments were tried, some with more success than others, but none of them were able to prevent many deaths. Despite the lethality of this disease, and the optimistic prospect of a vaccine, the stress that treating these patients places on health systems and the many unknowns about the exact pathology created by this virus contribute to making this disease the greatest medical challenge of recent times.
It is therefore interesting to see if all the tools that have been developed in network medicine for other diseases will help us to better understand COVID-19 and find better treatment options.
The published work presents CovMulNet19, a database containing all known and available interactions involving SARS-CoV-2 proteins, human proteins targeted by SARS-CoV-2, associated pathologies and symptoms as well as drugs that can potentially target them.
We used network analysis methods to identify diseases with similarity to COVID-19 and potentially effective drugs.
Our results show that the diseases most similar to COVID-19 are among the predispositions that have been reported as risk factors for severe forms of COVID-19, including intestinal, liver, neurological and other respiratory conditions.
This study demonstrates that CovMulNet19 can be used as an exploratory tool for therapeutic targeting or drug repositioning by analysing human proteins targeted by SARS-CoV-2, their interactions, and associated pathologies and symptoms.
Collaborations and acknowledgements
This study is a collaboration with Manlio De Domenico and Giuseppe Jurman of the Bruno Kessler Foundation in Trento, Italy.
The authors would like to thank INSERM, the Toulouse Cancer Santé Foundation and the Pierre Fabre Research Institute as Chair of Bioinformatics in Oncology of the CRCT as well as the BioInfo4Women programme of the Barcelona Supercomputing Center.
One picture
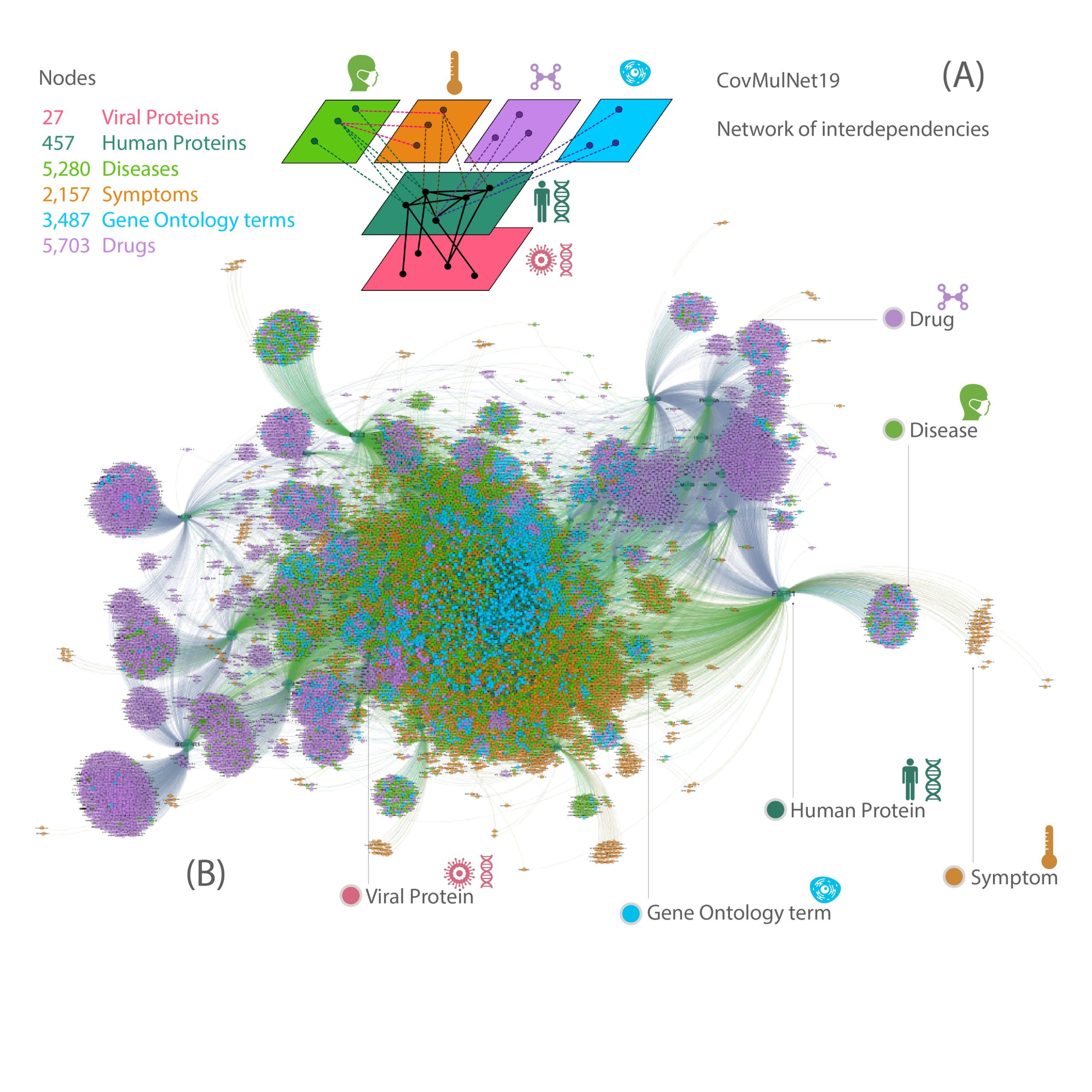 CovMulNet19: COVID-19 genotype-phenotype-drug interaction network. (A) Description of the different types of nodes and interdependencies between the different layers of the network corresponding to diseases, symptoms, drugs, GO (Gene Ontology) terms, human proteins and viral proteins. (B) Map of structural interactions (e.g. protein-protein) and functional interdependencies (e.g. protein-disease, protein-GO term, or disease-symptom). In total, the interaction network includes 1999 protein-protein links, 19,755 protein-disease links, 10,152 protein-symptom links, 13,018 drug-protein links, 9,210 protein-GO term links and 3,056 disease-symptom links. https://covmulnet19.fbk.eu
CovMulNet19: COVID-19 genotype-phenotype-drug interaction network. (A) Description of the different types of nodes and interdependencies between the different layers of the network corresponding to diseases, symptoms, drugs, GO (Gene Ontology) terms, human proteins and viral proteins. (B) Map of structural interactions (e.g. protein-protein) and functional interdependencies (e.g. protein-disease, protein-GO term, or disease-symptom). In total, the interaction network includes 1999 protein-protein links, 19,755 protein-disease links, 10,152 protein-symptom links, 13,018 drug-protein links, 9,210 protein-GO term links and 3,056 disease-symptom links. https://covmulnet19.fbk.eu
Discover the published article
Netw Syst Med. 2020 Nov 17;3(1):130-141.doi: 10.1089/nsm.2020.0011. eCollection 2020.
CovMulNet19, Integrating Proteins, Diseases, Drugs, and Symptoms: A Network Medicine Approach to COVID-19
Nina Verstraete, Giuseppe Jurman, Giulia Bertagnolli, Arsham Ghavasieh, Vera Pancaldi, Manlio De Domenico.

Toulouse Cancer Research Center (Oncopole)
Toulouse – FR
Contact us
+33 5 82 74 15 75
Want to join
the CRCT team ?




