
LEIMA KHAJAVI
ASSOCIATE INVESTIGATORS : JACOBO SOLORZANO, ALEXIS HUCTEAU, MIGUEL MADRID-MENCIA, ALEXIS COULLOMB & NINA VERSTRAETE
Deconvolution: description of the composition of the tumour microenvironment, towards personalised medicine
The increase in knowledge about the classification of different cancers and their treatments allows us to move towards personalized medicine, where each patient can receive a different treatment that truly corresponds to him/her. Our team is focused on the discovery of new targets related to the tumor environment that can be exploited for therapeutic purposes. We are also exploring the creation of artificial intelligence algorithms that can predict a patient’s response to a given treatment based on their medical history and clinical data.
To this end, our team is collaborating with numerous clinical and industrial partners to discover new therapeutic approaches for lung cancer patients. Determining the cellular composition of a sample is a priority for oncology research.
Building on the large amount of data available in the public scientific domain, our team is demonstrating innovative approaches to integrate these omics data to generate new and better signatures for supervised deconvolution algorithms.
This is a computational technique to determine the cellular composition of a (tumor) sample, in order to predict the response to immunotherapy treatments by using AI models.
Our main collaboration in this field is based on the LungPredict project (LungPredict: improve and personalize immuno-therapies – CRCT). Using deconvolution analysis, among other things, the goal is to extract and exploit the genetic information present in tumor samples from lung cancer patients. The data we obtain allows us to search for proteins and genes that may be potential therapeutic targets for personalized medicine approaches.
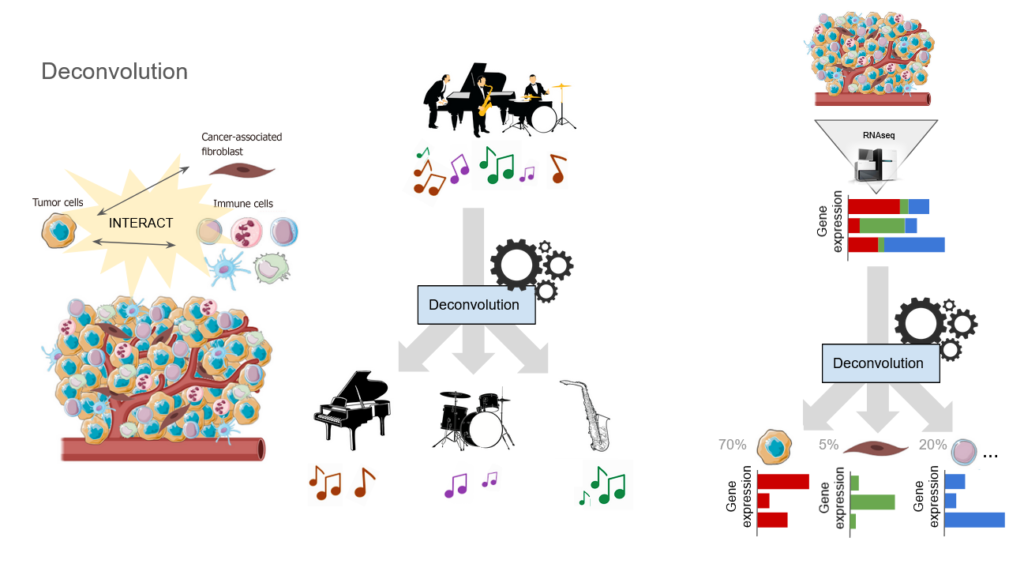
Deconvolution is a computational method to identify the composition of mixed samples from bulk measurements (such as gene expression or DNA methylation) based on the known profiles of specific cell types. We use it to identify the difference cell types that are present in tumour samples.
Github : https://github.com/VeraPancaldiLab/GEMDeCan
CRCT Chair of Informatics in Oncology (Fondation Toulouse Cancer Sante, Inserm et Pierre Fabre)

