A computational model of the formation of cells limiting the effect of promising cancer therapies
Cancer,
Computational bioinformatics,
systems biology;
Health sciences;
Immunology
mathematical models for health.
Many cancers can be treated by reinvigorating lymphocytes, our own bodies’ defenses against attacks, using specific drugs known as immune checkpoint blockers. Sadly, only a minority of patients and only some types of cancers can be treated this way. One of the reasons is that in some cases the cancer cells are able to corrupt other kinds of immune cells to protect themselves and prevent the infiltration of cancer-killing lymphocytes into the tumour tissue. In blood cancers, like chronic lymphocytic leukemia, it is known that just cultivating cancer cells with monocytes makes the latter turn into so-called nurse-like cells, which protect cancer cells, as the name suggests, from dying in the culture. If we could better understand how these cells are formed we would stand a chance to prevent their formation in patients and improve the success of novel therapies.
We used a computational model that represents cells as agents with specific behaviors to generate a simulation of interactions between monocytes and cancer cells in a culture dish. By examining what happens in the simulation, after checking that the simulation behaves like reality in a few situations, we could better understand what are the important processes that take place leading to the formation of the cancer-protecting cells. Also, we could see that cancer cells from different patients behaved differently and we hope that in the future our models will be able to make patient-specific predictions.
This work was a basic science contribution, but better understanding which processes and factors contribute to the formation of these cancer protecting macrophages could help us understand how to prevent these processes and have important consequences for patients treated with these therapies, which are becoming standard in an increasing number of cancers.
An important aspect of computational research is that we make the tools needed for these simulations publicly available so that anyone can reuse them, improve them and adapt them to different biological questions. In particular, we hope that this model will be a step towards establishing patient-specific simulations for personalized medicine.
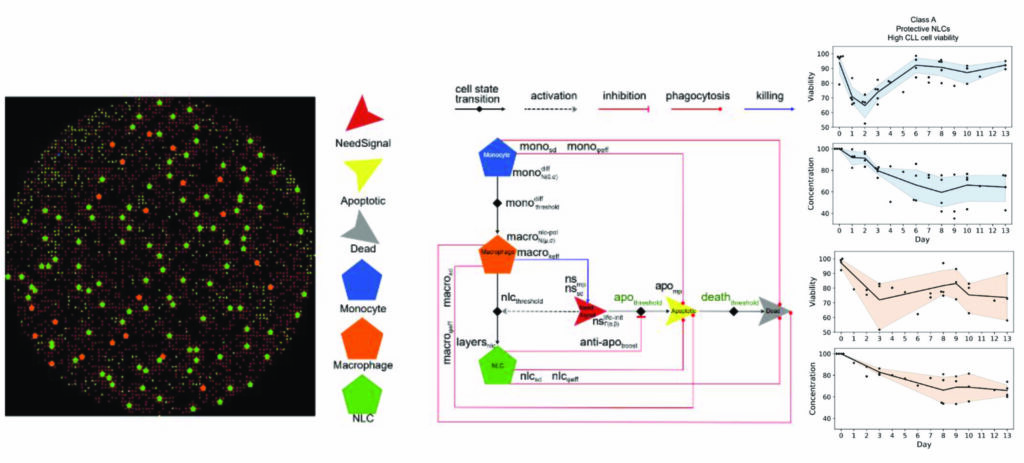
We designed a computational model of interactions between monocytes and leukemic cells that can be simulated to reproduce the dynamic of cell viability as observed in the experiments. The figure shows on the left an instant of the simulated culture dish, in the center the rules that determine intercellular interactions and on the right the predicted population dynamics compared to the data points obtained experimentally.
Discover the published article.
iScience 2023 May 19;26(6):106897.doi: 10.1016/j.isci.2023.106897. eCollection 2023 Jun 16.
An agent-based model of monocyte differentiation into tumour-associated macrophages in chronic lymphocytic leukemia
Nina Verstraete, Malvina Marku, Marcin Domagala, Hélène Arduin, Julie Bordenave, Jean-Jacques Fournié, Loïc Ysebaert, Mary Poupot, Vera Pancaldi
Collaborations and acknowledgements
Thanks to Nina Verstraete and Malvina Marku, Loïc Ysebaert, Mary Poupot
This study was funded by the Toulouse Cancer Santé Foundation and the Institut de Recherche Pierre Fabre as part of the CRCT Bioinformatics Chair in Oncology.

Toulouse Cancer Research Center (Oncopole)
Toulouse - FR
Contact us
+33 5 82 74 15 75
Want to join
the CRCT team ?




