Transcriptional DNA breaks and human disease
Olivier Sordet
Our research interests
The aims of our research are to understand how transcription, which is an essential cellular process, can threaten genome integrity in the context of DNA topological problems and to learn about its impact on the etiology and treatment of human disease. As they transcribe, RNA polymerases induce DNA torsional stresses whose defective removal can promote the formation of DNA breaks. The emerging picture is that transcription-associated DNA breaks are closely related to the formation of secondary DNA structures, such as R-loops and G-quadruplexes (G4s) (Figure 1). However, the interplay between transcription, DNA torsional stress and secondary DNA structures in the formation of DNA breaks is still puzzling. Our team aims to understand these connections to provide a better picture of how transcription threatens genome integrity and its link to human disease.
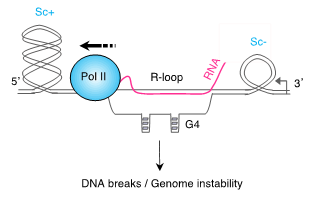
Figure 1. Connexions entre la transcription, la torsion de l’ADN (Sc : super-enroulement de l’ADN) et les structures secondaires de l’ADN (R-loop, G4) dans la formation de cassures de l’ADN / Figure 1. Interplay between transcription, DNA torsional stress (Sc: DNA supercoiling) and secondary DNA structures (R-loop, G4) in the formation of DNA breaks.
Questions raised
1. How does transcription induce DNA breaks in the context of DNA topology and secondary DNA structures?
Among genomic lesions, DNA double-strand breaks (DSBs) are infrequent but among the most harmful ones. Transcription-associated DSBs are particularly risky because they occur in transcribed regions, and thus potentially on disease-related genes, in both dividing and non-dividing cells. Our previous work identified a mechanism of DSB formation in non-replicating cells that strictly depends on transcription and which relies on both DNA topological constraints and R-loops (Sordet et al, EMBO Rep 2009; Cristini et al, Nucleic Acids Res 2016, Cell Rep 2019). In human cancer cells, we have further reported, in collaboration with Giovanni Capranico (Bologna, Italy), the interplay between R-loops and G4s in DSB induction (De Magis et al, PNAS 2019). Our goal is to characterize the extend of connections between DNA torsional stress and secondary DNA structures in the formation and repair of transcription-associated DNA breaks.
2. What is the contribution of transcriptional DNA breaks in human disease?
As we better understand how transcription can compromise genome integrity, it appears that the link between transcription and the etiology/development of human disease is much more important than previously anticipated (see our recent reviews: Cristini et al, Mol Cell Oncol 2020, Int Rev Cell Mol Biol 2021). For example, we reported that several genetic defects enhancing transcriptional DSBs also cause neurological disorders (Cristini et al, Cell Rep 2019). We are now focusing more specifically on transcription in promoting genome instability of cancer cells and its impact on the development and treatment of the disease.
Method used
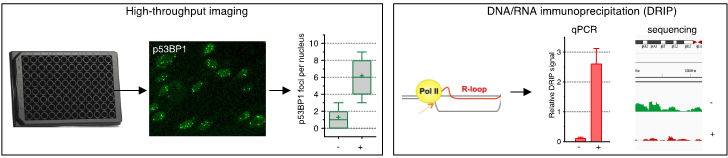
We use replicating and non-replicating cell models coupled with cutting-edge techniques to analyze transcription, DNA secondary structures and DNA damage at the cell and genome level. These techniques include high-throughput imaging, ChIP and DRIP (Figure 2). We also use the CRISPR technology to introduce in our cell models genetic defects found in human disease.
Keywords:
- Cancer;
- DNA break;
- DNA topology;
- Genome instability;
- G-quadruplexes;
- Human disease;
- Oncogenic translocation;
- R-loop;
- RNA polymerase;
- RNA/DNA hybrid;
- Topoisomerase;
- Transcription.
Group members
- Agnese Cristini (Ph.D.), Post-doc
- Mathéa Géraud (Pharm. D.), PhD student
- Lara Fernandez-Martinez, PhD student
- Latré Eustachia Lawson-body, Master2 student
Other team members involved
- Anne Pradines (Researcher)
- Julien Mazières (PU, PH)
- Estelle Taranchon-Clermont (Engineer)
Main external collaborations
- Giovanni Capranico, University of Bologna, Italy
- Béatrice Eymin, University of Grenoble, France
- Yves Pommier, NCI/NIH, Bethesda, USA
Selected publications
Int Rev Cell Mol Biol. 2021 Sep 12;364:195-240.doi: 10.1016/bs.ircmb.2021.05.001. PMID: 34507784.
Cristini A*, Géraud M, Sordet O*.
Transcription-associated DNA breaks and cancer: A matter of DNA topology.
*Co-corresponding authors.
Cell Rep. 2019 Sep 17;28(12):3167-3181.e6. doi: 10.1016/j.celrep.2019.08.041. PMID: 31533039
Cristini A, Ricci G, Britton S, Salimbeni S, Huang SN, Marinello J, Calsou P, Pommier Y, Favre G, Capranico G, Gromak N*, Sordet O*. Dual Processing of R-Loops and Topoisomerase I Induces Transcription-Dependent DNA Double-Strand Breaks.
*Co-corresponding authors.
Proc Natl Acad Sci USA. 2019 Jan 15;116(3): 816-825. doi: 10.1073/pnas.1810409116. PMID: 30591567
De Magis A, Manzo SG, Russo M, Marinello, Morigi R, Sordet O, Capranico G.
DNA damage and genome instability by G-quadruplex ligands are mediated by R-loops in human cancer cells.
Nucleic Acids Res. 2016 Feb 18;44(3):1161-78. doi: 10.1093/nar/gkv1196. PMID: 26578593
Cristini A, Park JH, Capranico G, Legube G, Favre G, Sordet O.
DNA-PK triggers histone ubiquitination and signaling in response to DNA double-strand breaks produced during the repair of transcription-blocking topoisomerase I lesions.
EMBO Rep. 2009 Aug;10(8):887-93. doi: 10.1038/embor.2009.97. PMID: 19557000
Sordet O, Redon CE, Guirouilh-Barbat J, Smith S, Solier S, Douarre C, Conti C, Nakamura AJ, Das BB, Nicolas E, Kohn KW, Bonner WM, Pommier Y.
Ataxia telangiectasia mutated activation by transcription- and topoisomerase I-induced DNA double-strand breaks.