
MALVINA MARKU
ASSOCIATE INVESTIGATORS: Nina Verstraete, Flavien Raynal, Miguel Madrid-Mencia
Boolean Regulatory Networks Of Cellular Phenotypes
In any given environment, the cellular processes that determine a cell’s phenotype consist in a cascade of gene interactions. Using the network representation, we model these interactions as regulatory networks, in which nodes represent the molecular entities while the connections represent the type and direction of interactions of different types. The experimental facilities in our lab, combined with advanced sequencing technologies and high-throughput analysis provide us important information about the expression of cell-specific genes under different extracellular conditions. Our goal is to use this information to infer the regulatory pathways involved in determining the cell phenotype, and build a dynamical model that would explain the temporal behaviour of the cellular system in various conditions. We used this approach to build a discrete (Boolean) dynamical model of macrophage differentiation into pro- and anti-inflammatory macrophages and, importantly, to identify the molecular pathways involved in the TAMs formation (Marku et al, 2020). A similar approach will be used to study other cells’ polarization spectrum and phenotype transitions.
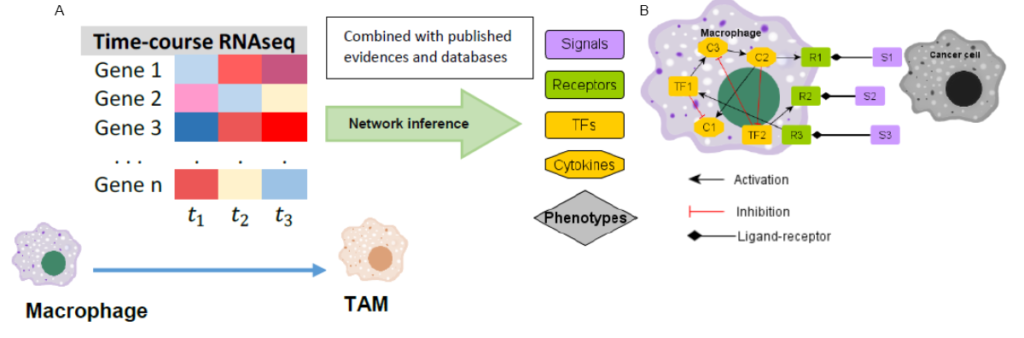
The dynamics of the processes taking place within the tumor will be studied at two levels: the intracellular level and the intercellular level. At the intracellular level, the phenotypic state and behavior of the cell will be determined by the activation of different pathways represented as gene regulatory networks (GRNs) in which a number of regulators determine the characteristics and behavior of the cell. We will apply mathematical methods to gene expression data over time (Figure A) to identify genes involved in specific cellular processes and derive regulatory models.
These intracellular regulatory patterns will then be integrated into models of interactions of the different cells in the samples (tumor sample or in-vitro) so that the behavior of each cell will be determined both by internal regulation and by interactions with other cells in the tumor microenvironment (Figure B).
Marku et al. 2020 https://www.mdpi.com/2072-6694/12/12/3664
CRCT Chair of Informatics in Oncology (Fondation Toulouse Cancer Sante, Inserm et Pierre Fabre)

