Équipe
Lorenzo Scipioni
OncoBreast :
Microscopie avancée pour l’étude de la chimiorésistance et de l’hétérogénéité dans le cancer du sein
Les spécificités
de notre axe de recherche
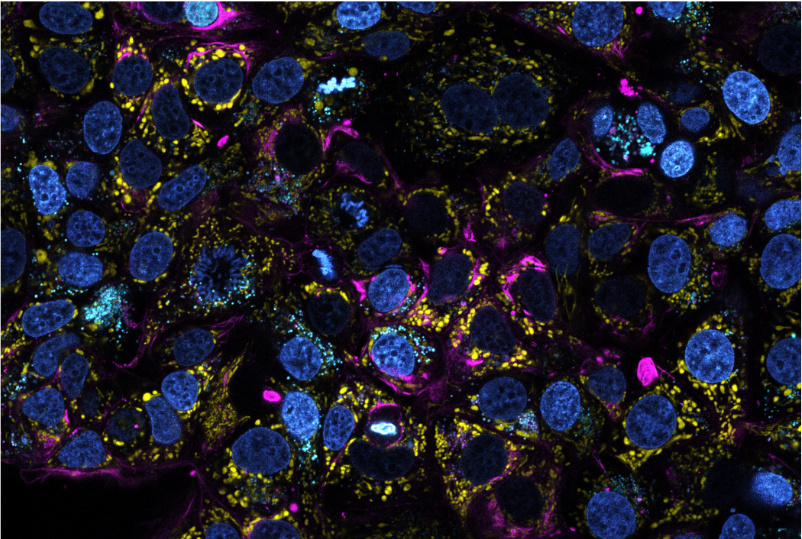
Les cellules, comme les êtres humains, possèdent de minuscules « organes », appelés organites, dont les rôles précis sont essentiels au maintien de la fonction cellulaire et déterminent l’aptitude de la cellule à accomplir des tâches spécifiques. Les propriétés des organites varient énormément d’un type de cellule à l’autre (par exemple, les cellules non tumorales et les cellules cancéreuses) et peuvent être utilisées pour « diagnostiquer » l’état physiologique d’une cellule et la manière dont elle peut réagir à des stimuli externes, tels qu’un traitement chimiothérapeutique.
Grâce à ESPRESSO (Environmental Sensors Phenotyping RElayed by Subcellular Structures and Organelles), une technique que nous développons et qui combine la microscopie avancée, les outils chimiques et l’informatique, nous sommes en mesure de suivre l’état physiologique de chaque cellule cancéreuse pendant de longues périodes, en caractérisant précisément les changements qui conduisent à la génération de populations chimiorésistantes.
Notre objectif principal est de poursuivre le développement de cette technologie révolutionnaire et de l’appliquer à l’étude du cancer du sein triple négatif, la forme de cancer du sein la plus agressive et la plus résistante aux thérapies. Dans ce contexte, nous prévoyons d’utiliser ESPRESSO comme outil de diagnostic et de pronostic pour améliorer la classification du cancer du sein triple négatif, la détection précoce des rechutes et le choix du traitement.
L’équipe Oncobreast combine les compétences et l’expertise de la physique, de l’ingénierie optique, de l’informatique, de la biologie et de la médecine, pour unir ses forces dans la lutte contre le cancer du sein.
Cancer du sein
Chimiorésistance
Rechute
Triple négatif
Microscopie
Hyperspectrale
Cellule unique
Spatiotemporelle
Protéomique spatiale
Organites
Apprentissage automatique
Réseaux neuronaux
DES PROJETS
DE RECHERCHE
Atlas des organelles du cancer du sein : combinaison d’ESPRESSO et de la protéomique spatiale pour étudier l’hétérogénéité des tumeurs
Lorenzo Scipioni & Giulia Tedeschi
Microscopie ESPRESSO : imagerie hyperspectrale plus rapide et plus profonde
Lorenzo Scipioni & Francesco Fersini
Propriétés des organelles en tant que marqueurs pour la détection précoce des rechutes dans les cancers du sein de type TNBC et Luminal B
Lorenzo Scipioni, Florence Dalenc & Camille Franchet
LES FOCUS
DE L’ÉQUIPE
Découvrir
Comprendre
Participer
PRODUCTIONS SCIENTIFIQUES
PUBLICATIONS 2026
PUBLICATIONS 2025
PUBLICATIONS 2024
LES MEMBRES DE L’ÉQUIPE
LES PARTENAIRES & FINANCIERS